About Iterative Screen Optimization
ISO was developed by the McLellan Lab at Dartmouth College, Hanover, NH (the lab has now relocated to the University of Texas at Austin, TX).
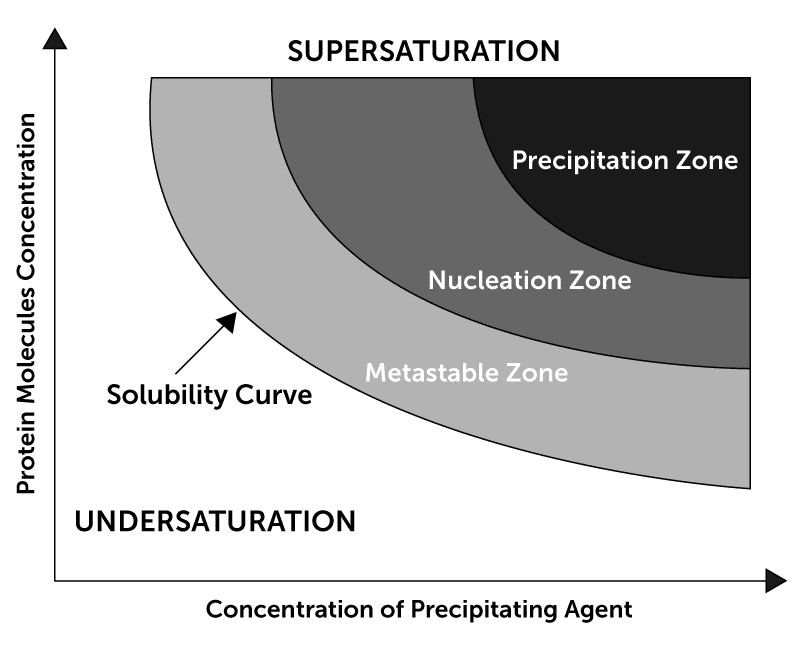
Iterative Screen Optimization (ISO) is a highly automated optimization process that can help researchers yield protein crystals from conditions that initially do not look promising.1
Using a premade screen, an initial screening experiment is set up. After images are collected on this experiment, a user classifies each of the images by scoring them as either Clear, Crystal, Light Precipitate, or Heavy Precipitate. Based on these scores, the ISO process creates a new experiment wherein the concentration of the precipitant from each condition is altered - either increased or decreased, to get into the nucleation zone in the protein solubility curve. For example, if the initial experiment yielded a clear score, the next ISO experiment will increase the precipitant concentration.
This ISO will then be performed on the newly created experiment (round 2) and such iterations will be done a few more times in a series. After the first iteration, every subsequent round of ISO takes into account the scores of the last two experiments to calculate the precipitant concentration in the next experiment. The ROCK MAKER® Crystallization Software can automatically generate experiments for each ISO step, adjusting the precipitant concentration of each condition based on user scores.
Reference
1. Jones HG, Wrapp D, Gilman MSA, Battles MB, Wang N, Sacerdote S, Chuang GY, Kwong PD, McLellan JS; Iterative screen optimization maximizes the efficiency of macromolecular crystallization; Acta Crystallogr F Struct Biol Commun.; 2019 Feb 1; 75(Pt 2): 123-131.
