About Iterative Screen Optimization
ISO was developed by the McLellan Lab at Dartmouth College, Hanover, NH (the lab has now relocated to the University of Texas at Austin, TX).
Iterative Screen Optimization (ISO) is a highly automated optimization process that can help researchers yield protein crystals from conditions that initially do not look promising.1
Using a premade screen, an initial screening experiment is set up. After images are collected on this experiment, a user classifies each of the images by scoring them as either Clear, Crystal, Light Precipitate, or Heavy Precipitate. Based on these scores, the ISO process creates a new experiment wherein the concentration of the precipitant from each condition is altered - either increased or decreased, to get into the nucleation zone in the protein solubility curve. For example, if the initial experiment yielded a clear score, the next ISO experiment will increase the precipitant concentration.
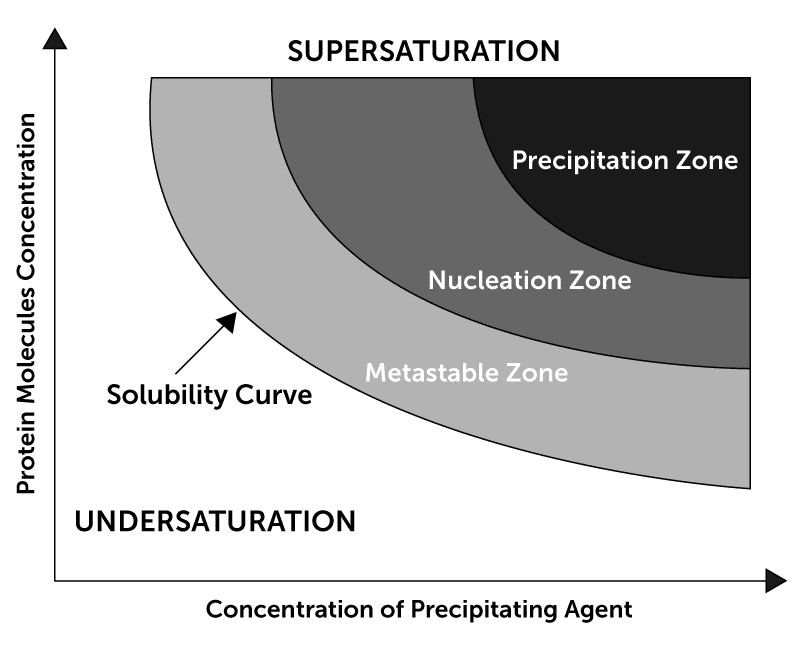
This ISO will then be performed on the newly created experiment (round 2) and such iterations will be done a few more times in a series. After the first iteration, every subsequent round of ISO takes into account the scores of the last two experiments to calculate the precipitant concentration in the next experiment. The ROCK MAKER® Crystallization Software can automatically generate experiments for each ISO step, adjusting the precipitant concentration of each condition based on user scores.
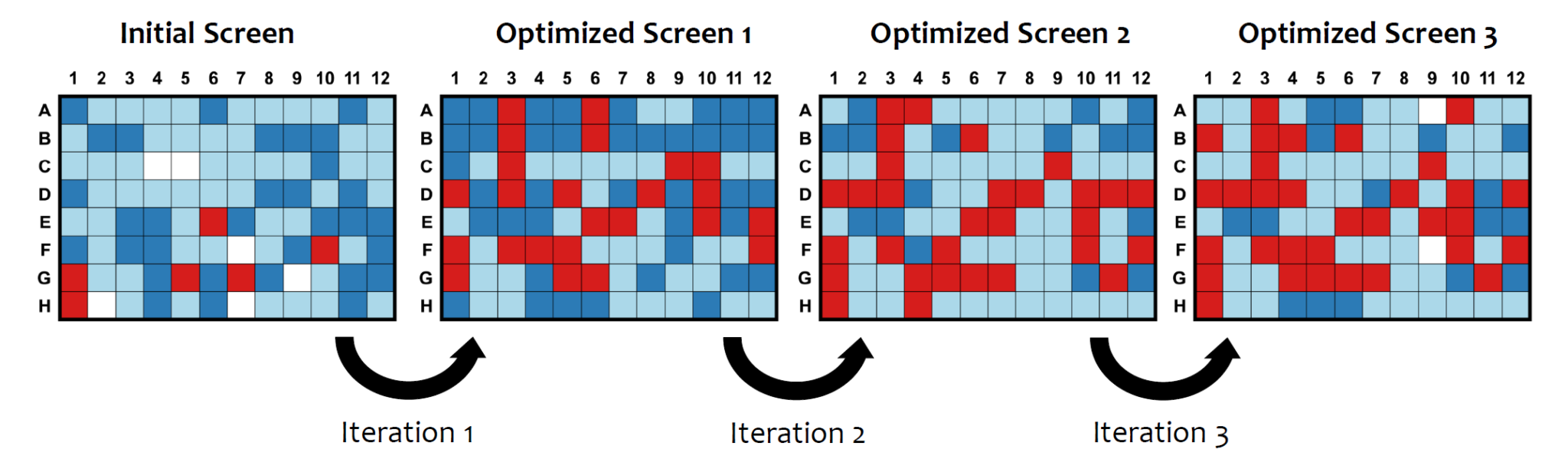
Example showing an increase in protein crystal hits (in red) with each iteration by using ISO strategy. Data from the McLellan Lab (UT Austin)
How ISO is Being Used to Grow Crystals
The Development of ISO at University of Texas at Austin

About Mr. Harrison Jones
Harrison earned a BS in Chemistry from the University of Utah in 2012. He worked at UCSD's Moores Cancer Center from 2012-2013 assisting in CLL clinical trials and characterizing novel pharmaceuticals. He joined the Dartmouth MD-PhD program in 2013 and his work primarily focuses on structural biology and biochemistry of respiratory viruses.
About the University of Texas at Austin
We are interested in elucidating the molecular mechanisms of host–pathogen interactions, particularly those involving viral glycoproteins. For this research, we exploit the synergy that exists between basic and translational science. Namely, the determination of structures and the development of tools needed to answer basic science questions can be translated into therapeutic interventions, which in turn can be used as reagents to further illuminate biological processes.
ISO Made Easy with ROCK MAKER® and FORMULATOR®
Implementing ISO into your crystallization research is fast and easy with our ROCK MAKER Crystallization Software and the FORMULATOR® Screen Builder. As ISO uses a 16-ingredient screen, the FORMULATOR is the only protein crystallization liquid handler capable of dispensing these complex ISO screens.
Simple ISO Workflow:
- Set up a new crystallization experiment
- Incubate and assign a score based upon visual inspection
- ROCK MAKER automatically adjusts the precipitant concentration of each condition based upon the assigned score
- Dispense optimized crystallization screen using the FORMULATOR Screen Builder
- Repeat steps 2-4 for each iteration
Reference
1. Jones HG, Wrapp D, Gilman MSA, Battles MB, Wang N, Sacerdote S, Chuang GY, Kwong PD, McLellan JS; Iterative screen optimization maximizes the efficiency of macromolecular crystallization; Acta Crystallogr F Struct Biol Commun.; 2019 Feb 1; 75(Pt 2): 123-131.
