Jeremy Lambert1, Tommy Duong2, Paul Scott1, Magnolia Bostick2, Andrew Farmer2
1FORMULATRIX, Inc., Bedford, MA 2Clontech Laboratories, Mountain View, CA
Miniaturization and scale-down of reagent and reaction volumes for low-input RNA-Seq workflows enables researchers to work with low sample input and increased numbers of samples. As the reaction volumes are reduced, accurate delivery of reagent volumes emerges as a critical step to maintain data quality and technical reproducibility. We present results from miniaturization of the Clontech SMART-Seq® v4 Ultra® Low Input RNA Kit for Sequencing and Illumina Nextera® XT workflows for generation of high-quality RNA-Seq samples using Mantis® , a compact desktop microfluidic dispensing technology.

Figure 3. Electropherograms demonstrating equivalent size distribution of cDNA products for full and small scale reactions.
Table 3. cDNA product yield (ng/ul) measured with PicoGreen assay.
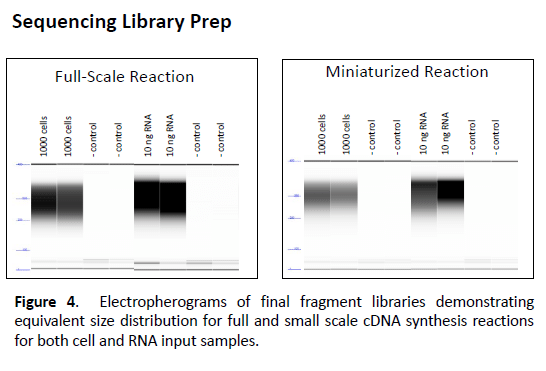
Figure 4. Electropherograms of final fragment libraries demonstrating equivalent size distribution for full and small scale cDNA synthesis reactions for both cell and RNA input ssamples
Reducing reagent volumes for cDNA synthesis using the Clontech SMART-Seq v4 Ultra Low Input RNA Kit for Sequencing provides results comparable to fullscale reaction volumes when using the Formulatrix® Mantis microfluidic dispensing system. Average yield of amplified cDNA from the scaled-down reaction volume workflow is compatible with input samples of cells and total RNA. RNA-Seq data analysis metrics are comparable between the full-scale and miniaturized reaction conditions. The combination of high-yield cDNA synthesis chemistry and high-precision low-volume reagent dispensing provides a workflow well-suited for large-scale RNA-Seq studies.
