Buffalo, NY
The Hauptman-Woodward Medical Research Institute’s High-Throughput Crystallization Screening Center (HTCSC) performs crystal-growth screening experiments using their proprietary 1,536 cocktail crystallization assay.
The HTCSC has been accepting samples from the scientific community for nearly 20 years and has enabled over 24 million crystallization experiments of >16,200 proteins. Initial crystallization conditions are identified for approximately half of these samples [Acta Cryst D64, 1123-1130 (2008) PMID: 19020350] supplied by hundreds of independent investigators.

How they do it...
Automated liquid-handling systems are used to dispense 1,536 crystallization cocktails and a protein solution into a single microassay plate previously filled with a protective layer of oil. It requires less than ten minutes and only 450 µL of protein to set up these experiments. Once complete, the plates are stored and imaged using a ROCK IMAGER 1000® equipped with SONICC® (Second Order Nonlinear Imaging of Chiral Crystals).
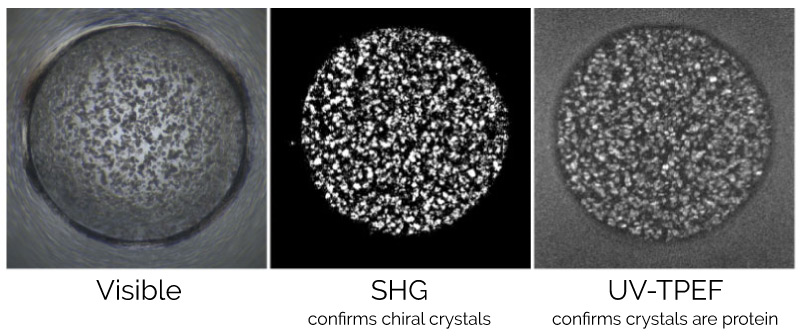
Three different “imaging parameters” are used: Visible, Second Harmonic Generation (SHG), and UV-Two Photon Excited Fluorescence (UV-TPEF). Each mode offers complementary information to help detect protein crystals and distinguish them from salt crystals.
Crystallization Method Development
The HTCSC currently uses the microbatch-under-oil crystallization technique, originally developed by Naomi Chayen, Patrick Shaw-Stewart and David Blow [J Appl Cryst 23, 297-302 (1990). J Cryst Growth 122, 176-180 (1992)] to set up crystallization experiments in high-throughput 1,536 well microassay plates [J Stuct Biol 142, 170-9 (2003) PMID: 12718929]. The HTCSC experiment identifies initial crystallization conditions, and aims to develop and improve crystallization technologies [Expert Opinion on Drug Discovery 6, 465-480 (2011) PMID: 22646073] including a 1,536 condition membrane protein crystallization screen [Protein Sci 18,1828-1839 (2009) PMID: 19554626], and methods to optimize and analyze screening results [Cryst Growth Des 11, 651-663 (2011) PMID: 21643490]. Most recently the HTCSC has been involved in a collaboration called the Machine Recognition of Crystallization Outcomes (MARCO), which uses machine learning algorithms to automatically analyze crystallization screening images [PLoS ONE 13, e0198883 (2018) PMID: 29924841]. The HTCSC is developing new methods for nanocrystal detection, as well as working in collaboration to generate novel crystallization cocktails and reagents.
Structures Solved
Since the HTCSC was established in February of 2000, they have aided researchers to solve the crystal structures of hundreds of soluble and membrane proteins. In 2016 alone, the HTCSC was cited in research publications for screening the conditions that led to the structure determination of ten soluble proteins and five membrane proteins. The use of automation and advanced imaging technologies has enabled the HTCSC to conduct these screening experiments quickly and increase the likelihood of crystal detection.
Click below to see a list of recent citations:
http://hwi.buffalo.edu/high-throughput-crystallization-center/structures-solved/
About the SONICC and UV-TPEF
The advanced imaging capabilities at the HTCSC significantly aids in the detection of protein crystals, especially micro- and nano-crystals that cannot be easily identified with traditional visible microscopy. SHG has an extremely low detection limit and can pick out <1μm crystals even in murky environments. The UV-TPEF modality helps distinguish between salt and proteinaceous material giving researchers the information they need in assigning hits. UV-TPEF and SHG are used routinely at the HTCSC to aid in crystal detection [Acta Cryst. F70, 993-1008 (2014)].
How to Use the HTCSC
To learn more about the scientific and technological expertise of the HTCSC investigators or to have your proteins set up for crystallization screening and imaging, please visit www.getacrystal.com or send an email to htslab@hwi.buffalo.edu.
