Accelerating Assay Development
When optimizing a particular process or assay, or moving an assay to large-scale production or screening, the design of an experiment is a crucial, but not always welcome, step. It is tempting to guess one’s way through it in order to minimize the use of both time and costly reagents. However, thoughtful assay development, also called Design of Experiments (DoE), ultimately leads to substantial cost and time savings. As examples, assays designed to quantify metabolite levels in blood, identifying and characterizing new drug molecules, setting up sensitive cell cytometry runs, and quantitatively measuring gene expression levels will all benefit from careful experimental design, which is also helpful when tweaking existing assays.
In drug discovery, an investigator must validate the assay methodology by proceeding through a series of steps along the pathway to high-throughput screening (HTS). The design of a typical HTS process might follow this line of reasoning:
Biology → Choice of assay format → Optimization of variables → Assay validation and adaptation → Assay optimization for HTS
The overall objective of any method validation procedure is to demonstrate that the method is acceptable for its intended purpose. The purpose can be to determine the biological and/or pharmacological activity of new chemical entities. The acceptability of a measurement procedure or bioassay method begins with its design and construction, which can significantly affect its performance and robustness.
One example of the optimization step is to determine the conditions that will optimize specific molecular interactions by systematically varying the concentrations of each component in the assay, as shown below:
| Factor | Concentration or condition |
| Enzyme | 0, 1, 10, 100 nM |
| Substrate | 0, 40, 400 nM |
| MgCl2 concentration | 0, 2.5, 5 uM |
| BSA | 0, 0.05%, 0.1% |
| Buffer type | Tris, HEPES |
| pH | 6, 7, 8 |
| Incubation time | 5, 10 min |
Ideally, this type of assay will combine every component at every concentration and condition, be carried out in duplicate or triplicate in 384- or 1536-well plates, and the reactions will be randomized over the plate to reduce the potential for errors, such as user error or plate effects. A common way of doing this is to use a software package such as JMP by SAS. This software will take all factors and conditions and construct a design customized to the problem, and then output a randomized list of all components specific for a given plate type. However, this list is exported as reagent concentrations, not volumes, meaning that the user will need to perform an additional step by creating a spreadsheet file to convert everything into volumes that can be dispensed. This step is both time consuming and creates a real potential for user error.
FORMULATRIX® assay development software, already included with both the MANTIS® and TEMPEST® liquid handlers, takes this troublesome step out of the process. With our software, investigators directly import the results from JMP, and the software automatically creates a dispense list that is ready to be used. It only requires a few steps: 1) Import JMP (or custom) data, 2) enter total well volume and stock concentrations, and 3) select the most appropriate chip for each reagent. The MANTIS or TEMPEST will then design a dispense list and, once the reagents are loaded on the system, the dispense can be run.
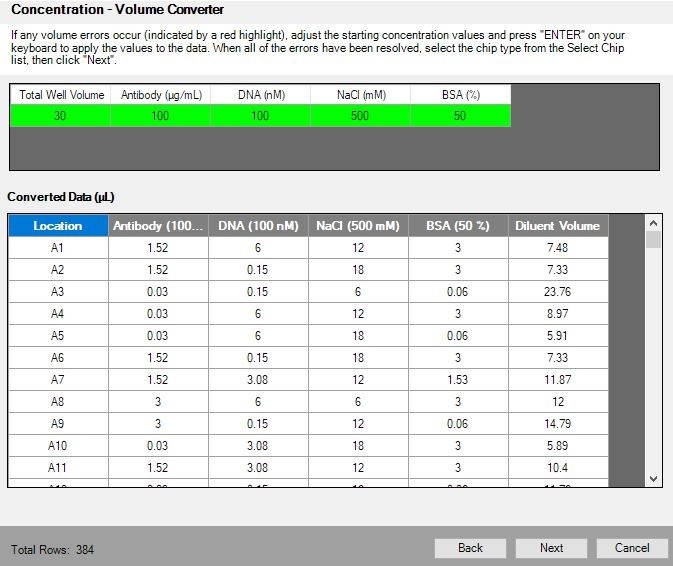
Shown above is a screenshot of the Volume Converter page of the assay development software, showing the exact volumes after conversion from concentration values. Here, investigators can modify stock concentrations to obtain volumes that will dispense fastest on their platform. On the next page, investigators select the most appropriate chip for each reagent, and the software will show how much, if any, rounding that was needed.
FORMULATRIX assay development software can greatly accelerate DoE by integrating almost seamlessly with JMP and removing common stumbling blocks in the process. Click below if you would like to see how it works and try it out in your own lab.
