Problem:
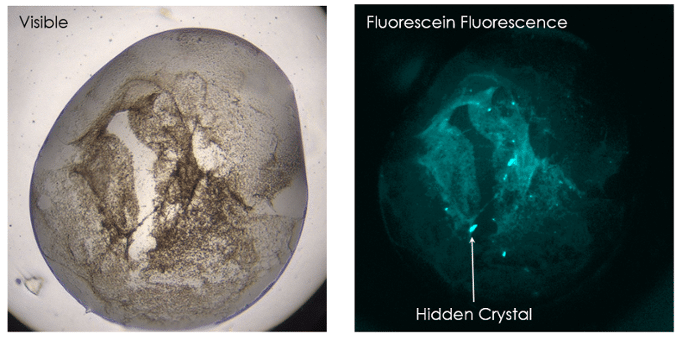
The unambiguous and reliable identification of biological crystals remains a major obstacle in crystallography, particularly in the critical stage of initial screening experiments. Automated imaging at high-resolution in the visible range is often insufficient to identify all conditions that have or may result in crystals.
Trace Florescence Labeling
One possibility to overcome the shortcomings of intrinsic fluorescence, which critically depends on the amount of naturally occurring tryptophan, is Trace Fluorescence Labeling (TFL). There are established protocols that include the rapid labeling of 0.1% of samples with a variety of fluorescent dyes immediately prior to crystallization. In addition to a significantly enhanced signal to noise ratio over intrinsic fluorescence, multiple labeling can be used to verify the presence of multi-subunit complexes in the crystal.
Sensitive Crystal Detection with MFI
The use of UV light and TFL imaging adds an alternative tool in crystal detection.
Multi-Fluorescence Imaging (MFI) provides the flexibility to image in 3 different wavelengths of the user’s choosing including UV and visible fluorescence to easily detect protein crystals, even those buried under precipitate.
With UV imaging, drops are illuminated with UV light and the fluorescence generated from aromatic amino acids like tryptophan are detected to create an image. By labeling only 0.1% of a sample with a fluorophore, TFL also provides the ability to detect proteins with little-to-no tryptophan.
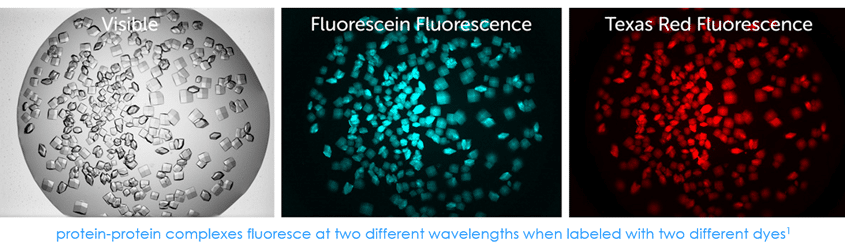
Find Protein-Protein Complexes
With Multi-Fluorescence Imaging (MFI), you can now differentiate between crystals of a protein-protein complex and crystals of just one protein. Simply, label the two suspected proteins, or subunits, with two different amine reactive dyes and then image your drop at the two corresponding wavelengths. Crystals that fluoresce at both wavelengths are protein-protein complexes, whereas those that fluoresce at only one wavelength are of a single protein.
Large Range of Magnifications
With 3 objectives, you can zoom out to see the entire drop or zoom in to see your crystals in greater detail. Digital zooming of the 9MP images allows you to match the FOV directly to your visible image.
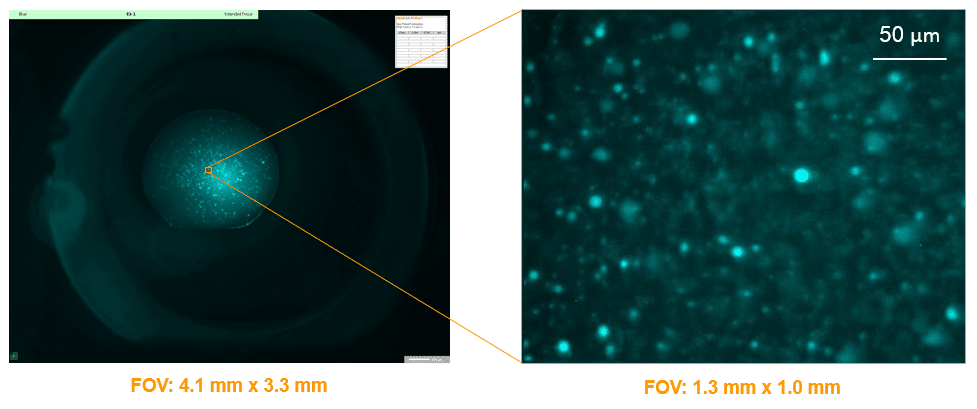
Image 3 Different Wavelengths
Choose up to 3 different light wavelengths to image your crystals, including ultraviolet and various visible light wavelengths. Interchangeable filter modules are used to maximize image quality and dye-label compatibility.
Up to three filter modules can be installed at one time and are easily switched out. The default filter configuration includes: UV fluorescence, Fluorescein, and Texas Red filter sets.
Quick + Safe Labeling that Lasts
Simple 0.1% protein labeling protocol
1.Prepare a 5mM stock solution of a succinimidyl ester dye in DMSO.
2.Add appropriate amount of dye to protein solution for 0.1% labeling of lysine residues assuming a 1:1 stoichiometric labeling efficiency to amine residues.
3.Wait 5 minutes at which point 90% of the dye is bound.
There is no need for purification and samples are still fluorescent after 120 days. Protein labeling with fluorophores has been shown to have no known negative impact on crystallization and demonstrated as a useful method for positively identifying protein crystallization hits.2,3,4,5
References
1 Images acquired at Maier Lab at Biozentrum Basel
2 Labeling protocol developed by Moritz Hunkeler, Maier Lab at Biozentrum Basel
3 Pusey M., et al. Acta Cryst. (2015) F71, 806-814
4 Watts, D., et al. Acta Cryst. (2010) D66, 901-908
5 Groves, M.R., et al. Acta Cryst. (2006) D63, 526-535
