Find Hidden Crystals
Discover crystals buried in precipitate or crystals not visible by the naked eye.
Don't Miss Hits
Detect thin crystals, microcrystals <1 μm, and crystals obscured in birefringent LCP.
Target the Right Crystals
Know if your detected crystals are protein or not using integrated UV imaging.
Remove the guesswork.
SONICC finds protein crystals that other technologies cannot.
The ultimate imaging technology in visualizing protein crystals, Second Order Nonlinear Imaging of Chiral Crystals (SONICC) definitively finds crystals buried in precipitate or submicron crystals naked to the eye. Two powerful technologies, Second Harmonic Generation (SHG) and Ultraviolet Two-Photon Excited Fluorescence (UV-TPEF) are combined together in a completely automated imager to quickly image your high throughput crystallization plates and positively identify protein crystals.
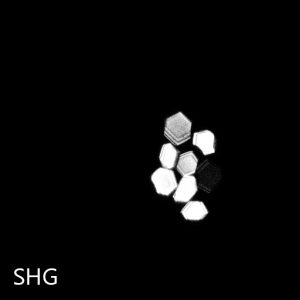
Crystals appear white against a stark black background, helping you to identify crystals even in murky environments. SONICC can detect extremely thin crystals, microcrystals <1 μm, and crystals obscured in birefringent LCP.
Major pharmaceutical and academic research labs worldwide find SONICC to be extremely successful, cost-effective, and efficient.
Find Hidden Crystals That You May Have Missed Before
The unique imaging properties of SONICC allow crystal detection in almost any optical environment, including opaque and turbid environments. Only chiral crystals, such as proteins, produce a signal using second harmonic generation (SHG). This imaging technology reveals very thin protein crystals or those buried under precipitant.
Detect Sub-Micron Crystals
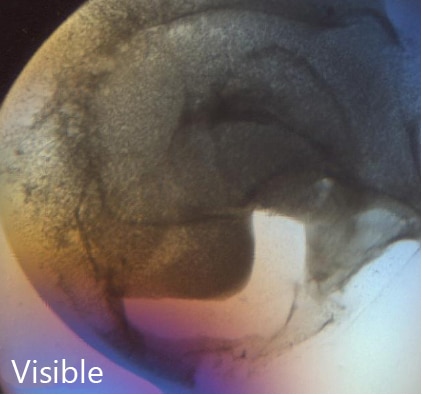
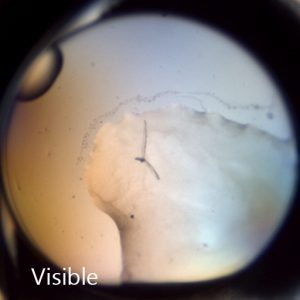
SONICC can detect nano-crystals with the use of high numerical aperture objectives. Small crystals that may be indistinguishable from precipitant can be clearly differentiated as shown in the image to the right. This positive hit indicates a condition for further optimization, which may have been missed without SONICC technology.
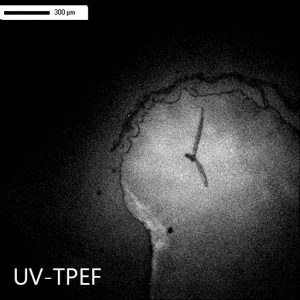
Distinguish Salt Crystals from Protein Crystals
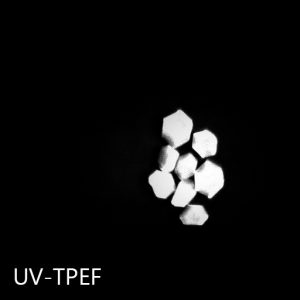
The UV-TPEF (Ultraviolet Two-Photon Excited Fluorescence) mode is analogous to traditional UV fluorescence and creates images based on the fluorescence of UV excited amino acids such as tryptophan.
UV-TPEF is a multiphoton imaging technique that uses longer wavelengths of excitation providing greater plate compatibility, less damage to your samples, and confocal imaging.
"SONICC SHG has been very useful for us in detecting initial crystallization conditions in LCP for membrane protein targets. These conditions usually contain very small crystals and would have been missed by UV imaging especially when the drop is clouded by aggregation or as a result of the opaque LCP matrix."
Quickly Identify Obscured Crystals
SONICC technology produces high contrast black and white images, making it easier to find hidden microcrystals. Below are 96-well plates imaged in visible light (left) and by SONICC in SHG mode (right). Each well-containing crystals is instantly apparent when imaged with SHG.
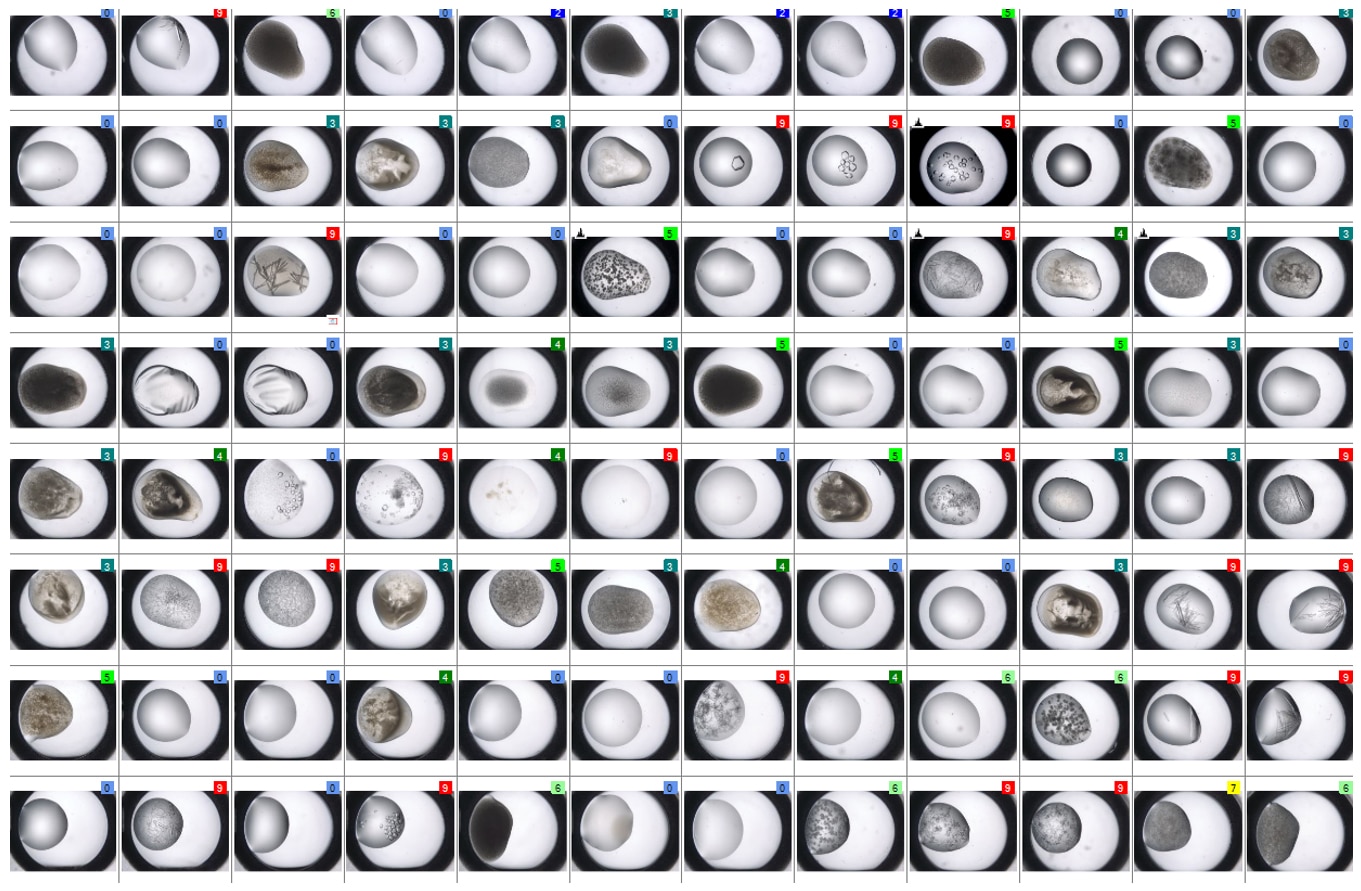
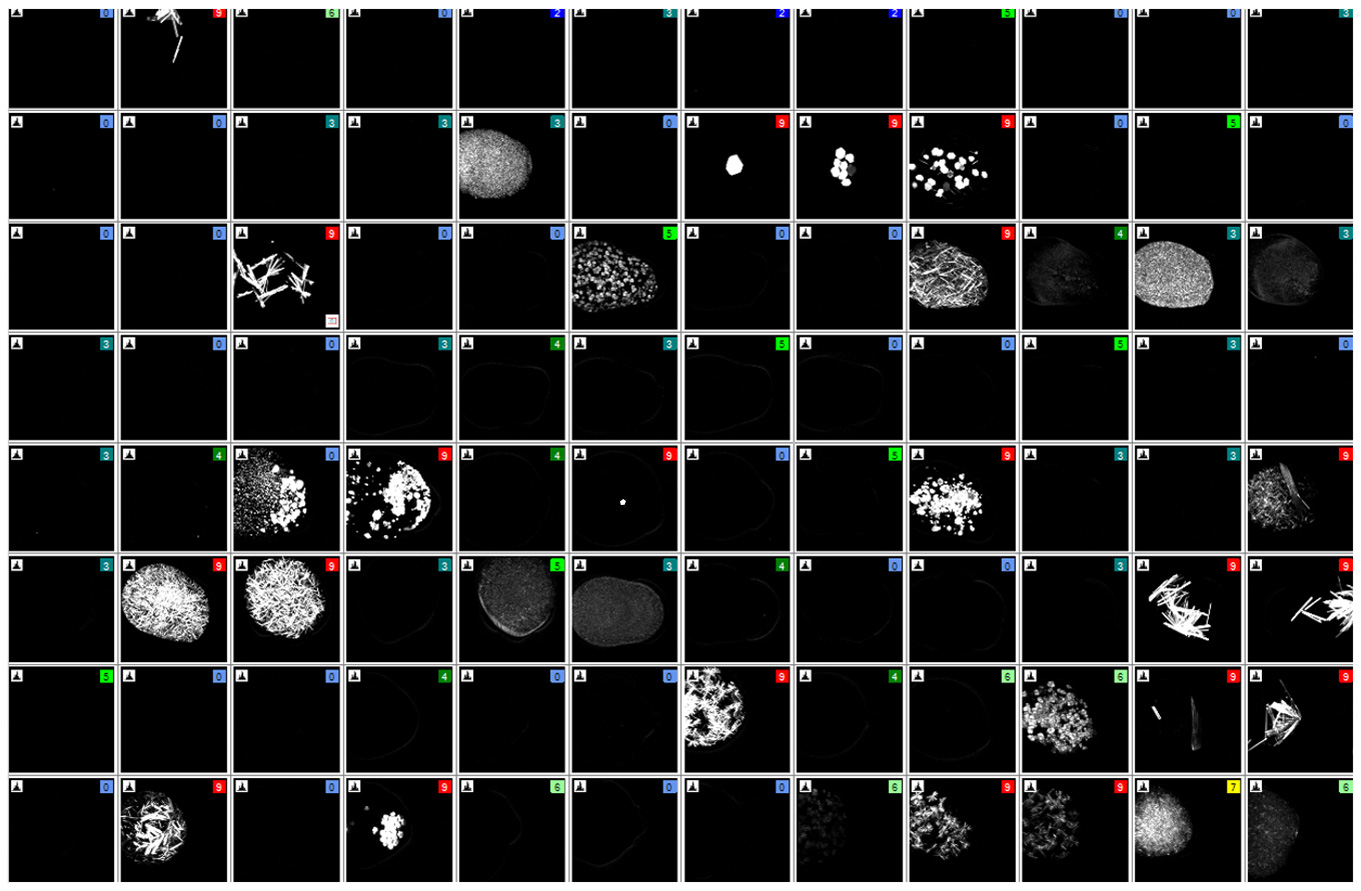
Drag the orange slider to quickly discover which wells have buried crystals detected with Second Harmonic Generation
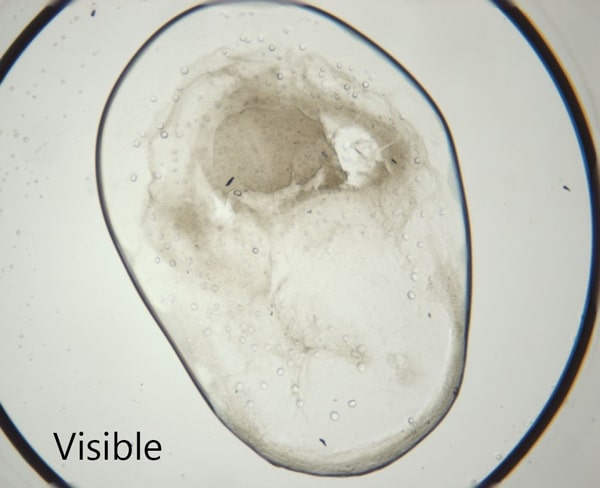
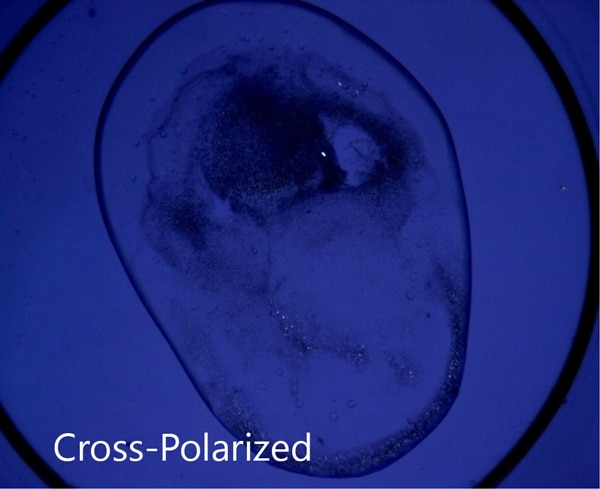
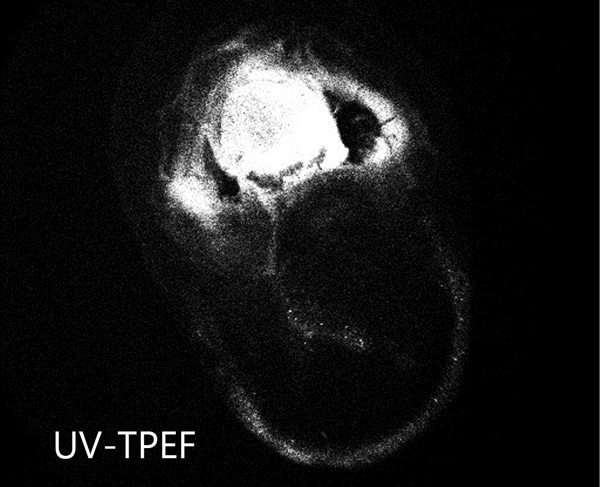
Ideal for Imaging in Lipidic Cubic Phase (LCP)
The extremely low detection limit of SONICC makes it the best imaging technology for LCP plates. Small crystals, buried in turbid lipidic cubic phase can easily be visualized with SONICC. Drop location and autofocus are both used to quickly and accurately find the LCP drop. The accompanied visible imaging techniques of crossed-polarized imaging offer complementary information all with extremely fast imaging times.
Models to Fit Your Workflow
SONICC comes in two configurations to better fit your lab's needs:
A benchtop model for imaging a single plate at a time. Best for lab's where throughput is not a concern.
An integrated model that is built into a ROCK IMAGER 1000 allows you to store and automatically image up to 940 plates. Two plates can be imaged at the same time - one with visible light and one with SONICC. This allows up to 480 96-well plates to be imaged in visible light and more than 96 plates in SHG or UV-TPEF in just one day.