In providing various next-generation sequencing (NGS) services relating to a wide range of customers, sample types, and sequencing approaches, core laboratories must balance the efficiency of their NGS workflow with the quality of the data that they generate. As the cost of sequencing declines, the expenses associated with library preparation begin to dominate the cost of the entire NGS workflow. In order to address this dominant expense in sequencing, many institutions are adopting the trend of reaction miniaturization, effectively reducing volumes to as little as 10% of the manufacturer's recommended volume.
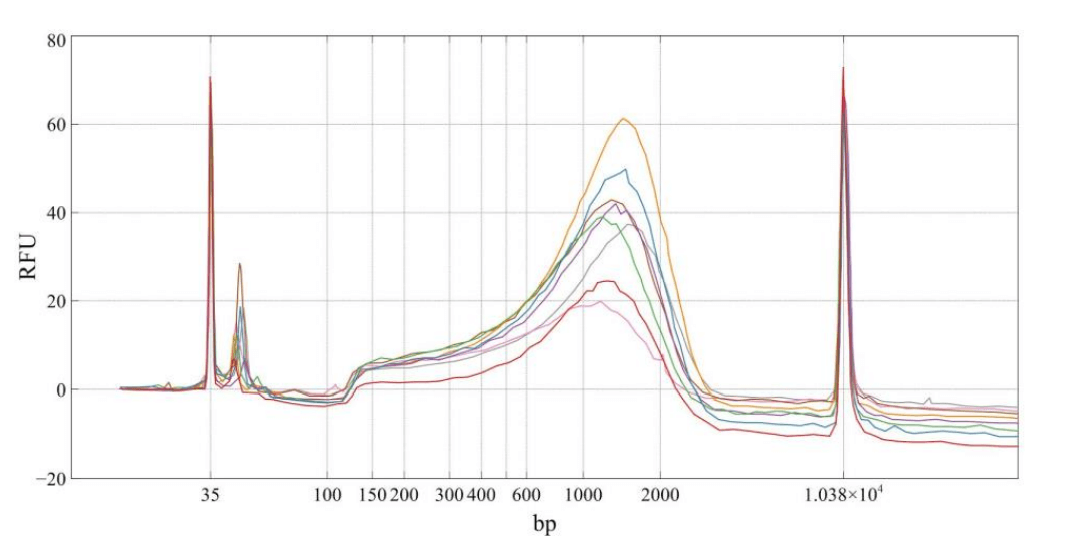
This application note highlights a workflow for generating high-quality NGS libraries, even from low cellular input, through precise microfluidic reagent dispensing. The quality control data for quarter volume Illumina Nextera XT library preparation showcases reaction miniaturization's applicability and efficacy in single-cell sequencing.
The MANTIS® Liquid Dispenser from facilitates reaction miniaturization through precise microfluidic reagent dispensing at volumes as low as 100 nL and is consumable-free, simple to use, and reliable.
