Protein crystallization is the process of forming a regular array of individual protein molecules stabilized by crystal contacts. Understanding the protein structure is of great importance for predicting its function, studying protein-protein or ligand-protein interaction for drug discovery, and uncovering stages in enzyme catalysis.
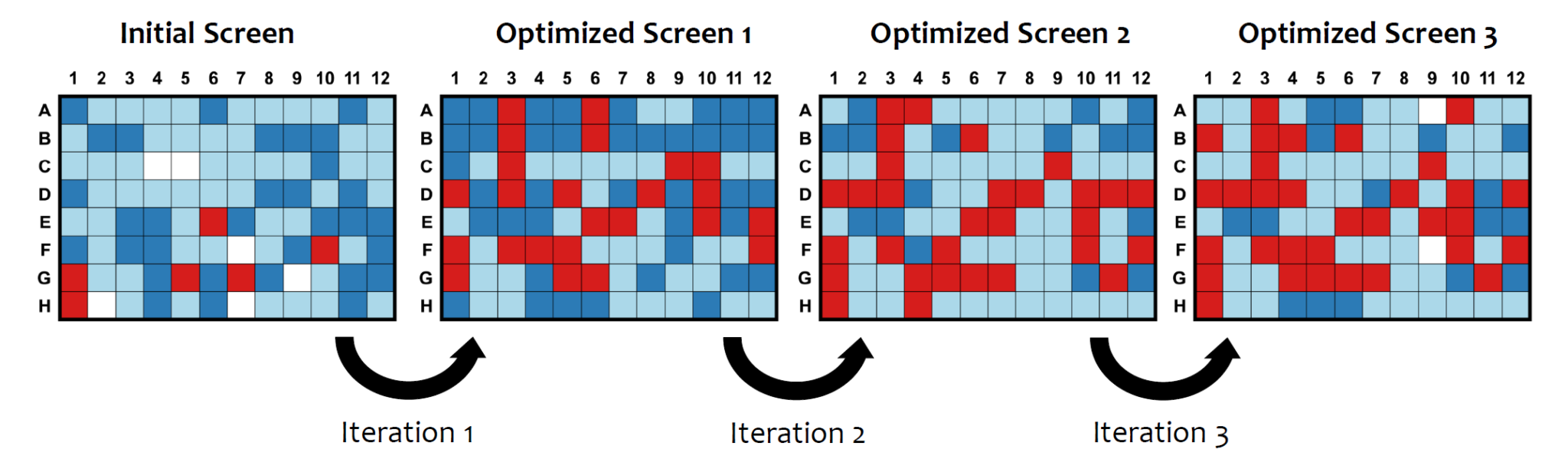
The process of protein crystallization involves gradually reducing the solubility of the protein using precipitants in a controlled environment (Figure 1). Predicting the optimal conditions under which a protein will form individual and well-diffracting crystals remains a challenge due to a complex interplay of the interactions between protein molecules.
Therefore, a comprehensive screening of various conditions, such as pH, temperature, salt concentration, precipitant type, and other additives, is necessary to optimize crystal development.
Figure 1: Protein Crystallization Phase Diagram
Protein Crystallization Workflow
Protein crystallization is a multistep process, beginning with the experimental design (Figure 2). Next, crystallization plates are prepared by combining purified protein droplets with the screening reagents. This is followed by incubation at specific temperatures and imaging to check for crystal formation.
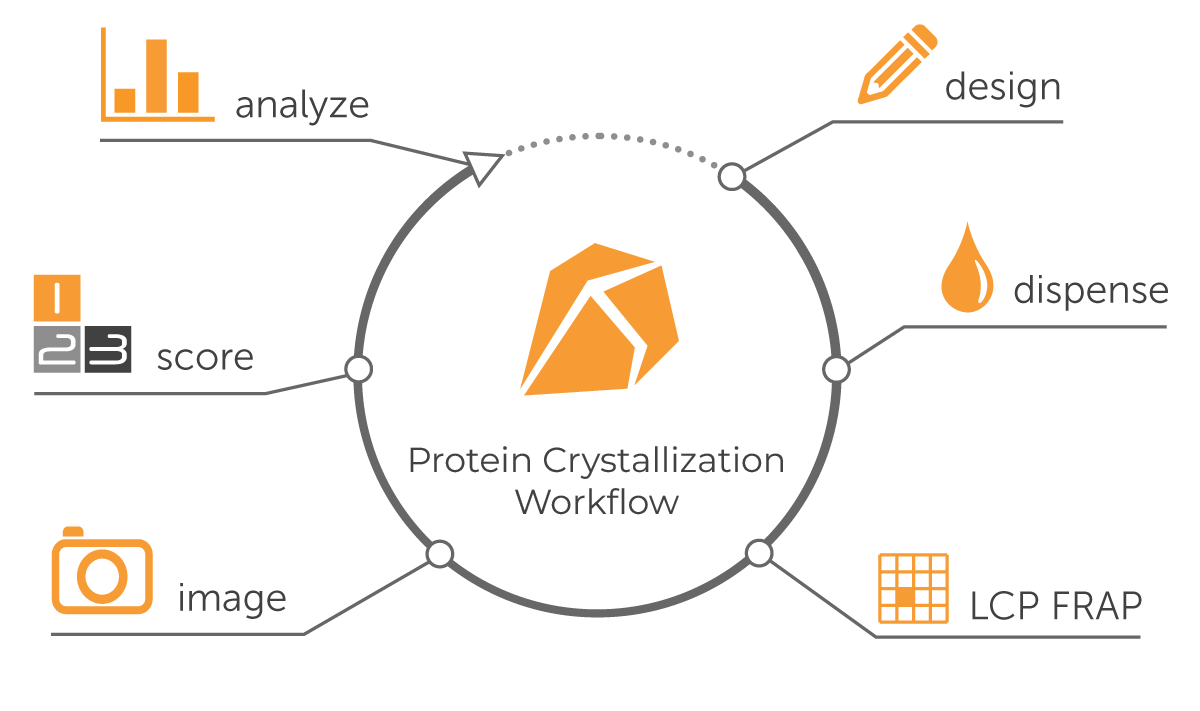
Figure 2: Protein Crystallization Workflow
Protein Crystallization Techniques
There exist various protein crystallization techniques, and the choice depends on multiple factors, including the type of protein, its concentration and available volume, required experimental throughput, type of reagents to be used, and desired outcomes. Some of the most widely used protein crystallization methods are given below:

| Feature | Hanging Drop | Sitting Drop | Micro-Batch | Micro-Dialysis | Free-Interface Diffusion | Lipid Cubic Phase |
|---|---|---|---|---|---|---|
| Amount of Protein | Small to Large | Small | Small | Large | Very Small | Small to Large |
| Automation | Possible | Possible | Not Possible | Not Possible | Difficult | Possible |
| Seeding | Possible | Possible | Not Possible | Not Possible | Possible | Possible |
| Harvesting | Easy | Easy | Difficult | Easy | Difficult | Difficult |
| Suitability | Crystallization Optimization Using High Surface Tension Reagents | Initial Screening | For Proteins and Reagents Having Minimal Interactions with Oil | For Developing Large Crystals | For Diffraction-Quality Crystals | For High-Quality Crystals of Membrane Proteins |
Table: Comparison of Protein Crystallization Techniques
Imaging Modalities
Different imaging modalities, including visible light, ultraviolet, multi-fluorescent imaging, and SONICC®, are available to image the protein drops and identify positive hits. Each technology has its own advantages; the choice varies based on the crystal properties and the purpose of imaging. A brief description of each modality is given below:
Visible Light Imaging
Visible light imaging captures images using the visible light spectrum (400 to 700 nm wavelength). Techniques such as bright-field, dark-field, and phase-contrast microscopy utilize visible light for imaging. This modality is suitable for analyzing large-sized protein crystals, but cannot distinguish between protein and salt crystals.
Ultraviolet (UV) Imaging
UV imaging is a label-free imaging modality where protein drops are illuminated with UV light. The fluorescence from aromatic amino acids like tryptophan helps distinguish protein crystals from salt ones. However, it may give false-positive results with phase separation and protein aggregation.
Multifluorescence Imaging (MFI)
This is a powerful technique for imaging crystals of fluorescently labeled proteins using trace fluorescent labeling approach (TFL). MFI efficiently distinguishes protein crystals from salts, as well as the crystals of a single protein from a complex. However, choosing the type of fluorescent dye and its concentration is important as it impacts the protein stability and, ultimately, its crystallizability.
Second Order Non-linear Imaging of Chiral Crystals (SONICC)
The SONICC combines Second Harmonic Generation (SHG) with Ultraviolet Two-Photon Excited Fluorescence (UV-TPEF) to image the protein crystals. It easily detects microcrystals (<1 μm), and the crystals obscured in birefringent LCP or buried under aggregates.
Need for Automation in Protein Crystallization
The conventional protein crystallization process has been slow, resource-intensive, and error-prone due to manual workflows for screen building, plate setup, and imaging. It often involves working with sub-microliter volumes, and even slight inaccuracy in dispensing or mixing the reagents leads to significant errors or suboptimal results. Automation and integration of all crystallization steps eliminate human error, ensure reproducibility, and dramatically increase the throughput.
Automation Solutions for Protein Crystallization by Formulatrix®
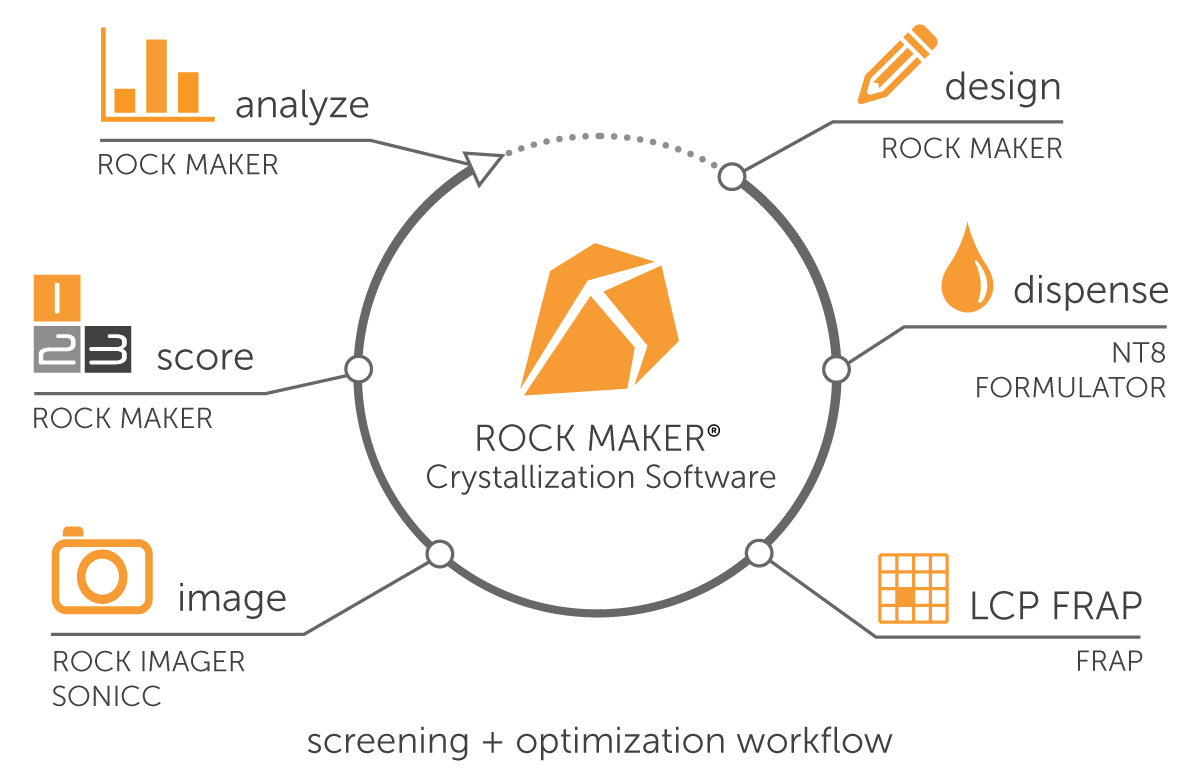
Formulatrix protein crystallization repository includes Laboratory Information Management Software (Rock Maker®), screen builder (Formulator®), drop setter (NT8®), and imagers (Rock Imagers®). Our systems are designed to streamline protein crystallization workflows with high precision, throughput, and time efficiency.
Rock Maker - Crystallization Software
Rock Maker is a powerful and easy-to-use protein crystallization software that seamlessly integrates all our protein crystallization products to manage the entire workflow, including data analysis.
Formulator - Screen Builder
The Formulator is a microfluidic liquid dispenser designed to set up crystallization screens and capable of dispensing up to 34 different ingredients of any volume and viscosity with a 96-nozzle chip. It supports low volumes (down to 200 nL) with no upper limit and can dispense a 100 μL, 3-ingredient grid across 96 wells in 2.7 minutes, accommodating all microplate types. Features include an integrated barcode scanner, automatic ingredient detection, and seamless Rock Maker crystallization software integration.
NT8 - Drop Setter / Crystallization Robot
The NT8 Drop Setter is a fast liquid handler capable of dispensing drops from 10 nL to 1.5 μL. It supports various experimental setups like hanging and sitting drops, LCP, additives, and seeding. It features proportionally controlled active humidification to prevent evaporation. Additionally, reusable tips and compatibility with temperature-controlled rooms ensure cost efficiency, flexibility, and reproducibility. Moreover, it is the only crystallization robot that integrates with Rock Maker.
Fluorescence Recovery After Photobleaching (FRAP) for Membrane Proteins
Membrane proteins are challenging to crystallize in LCP, and the optimization process is time, effort, and resource-intensive. FRAP saves all the hassle and resources by quickly screening LCP conditions and ruling out sub-optimal conditions before setting up crystallization experiments.
Rock Imager - Protein Crystallization Imagers
Rock Imagers are a series of automated imaging systems designed for protein crystallization screening. They capture superior-quality images of protein drops and are available with various plate capacities and imaging features (Table).
| Feature | Rock Imager 1 | Rock Imager 2 | Rock Imager 360 | Rock Imager 1000 |
|---|---|---|---|---|
| Plate Capacity | 1 | 2 | 360 | 1000 |
| Refrigeration | No | No | Yes | Yes |
| Plate Storage and Retrieval | No | No | Yes | Yes |
| Imaging Options | Visible, UV | Visible, UV, MFI | Visible, UV, MFI | Visible, UV, MFI, SONICC |
| Footprint | Benchtop | Benchtop | Benchtop | Floor Standing |
| Software | Windows-based | Windows-based | Browser-based | Windows-based |
AI-Based Autoscoring Models Integrated with Rock Maker
Integration of MARCO and Sherlock, AI-based autoscoring models, with Rock Maker has streamlined the analysis of the extensive image datasets generated during crystallization experiments. Sherlock, our proprietary autoscoring model, is continually being enhanced based on feedback from Rock Maker users to further improve its performance and accuracy.
Related Links
Webinars
Discover how Formulatrix’s comprehensive suite of tools enables a seamless, fully integrated automated protein crystallization workflow.
Learn how combining the advantages of the microfluidic format with high-throughput imaging opens exciting perspectives in protein crystallization studies.
Learn how to enhance and automate the scoring of crystallization experiments using Sherlock, an advanced AI auto-scoring model that can help you save time and increase your confidence in the results.
Explore the latest advancements in benchtop imaging technology offering unmatched plate storage, integrated refrigeration, and advanced automation for effortless crystal imaging.
Application Notes
Learn how our protein crystallization software- Rock Maker can help you perform effortless AI-based autoscoring of your crystallization experiments.
Learn how our protein crystallization software- Rock Maker can help you perform effortless AI-based autoscoring of your crystallization experiments.
Learn more about our advanced liquid handler-NT8 to set up crystallization experiments with reliable low volume performance due to its built-in active humidity control system.
Learn more about our advanced liquid handler-NT8 to set up crystallization experiments with reliable low volume performance due to its built-in active humidity control system.
Publications
FAQs
What is automated protein crystallization, and how does it enhance efficiency compared to manual methods?
Automated protein crystallization involves setting up crystallization experiments using instruments that streamline each step of the workflow, minimizing the need for manual intervention. By reducing pipetting errors, conserving valuable protein samples, and enabling high throughput under precisely controlled conditions, automation greatly enhances experimental efficiency. This allows faster screening, streamlined optimization, and more reliable results with less hands-on time.
How does automation reduce human error in protein crystallization experiments?
Automation eliminates inconsistencies in liquid handling and drop setup that occur with manual pipetting. By standardizing liquid dispensing, mixing, and environmental control, automated systems ensure consistent drop volumes and conditions across experiments. This precision reduces setup errors, improves reproducibility, and enhances the reliability of crystallization outcomes.









![[Updated 2025] ROCK MAKER V4 - 2 (1)](https://fmlx.b-cdn.net/wp-content/uploads/2025/07/Updated-2025-ROCK-MAKER-V4-2-1.jpg)