Gencove Implements Efficient and Reproducible Sequencing to Accelerate Genomic Discoveries

Gencove is paving the way to accessible and interpretable personalized DNA sequencing through utilization of Low-Pass Whole Genome Sequencing (LOW-PASS WGS). Unlike other personal genomics companies, Gencove sequences their customer's DNA, allowing them to uncover variants outside the scope of traditional Genotyping Arrays.

However, sequencing can be expensive, and in order to prepare thousands of NGS libraries cost effectively, Gencove worked with the Innovation lab at the New York Genome Center to identify the most efficient and reproducible automated sample preparation tools available. To our delight, the MANTIS® Liquid Handler was selected as the tool-of-choice for nucleic acid normalization in Gencove's high-throughput library preparation workflow!
To learn about Gencove's approach to personalized sequencing, the impact on precision medicine, and how Gencove achieves cost effective NGS library preparation, we spoke with Kaja Wasik, CSO and Co-founder in this Customer Spotlight from FORMULATRIX®.
FORMULATRIX® (F): How is Gencove paving the way to accessible and interpretable personalized DNA sequencing, and how does this fit into the future of precision medicine?
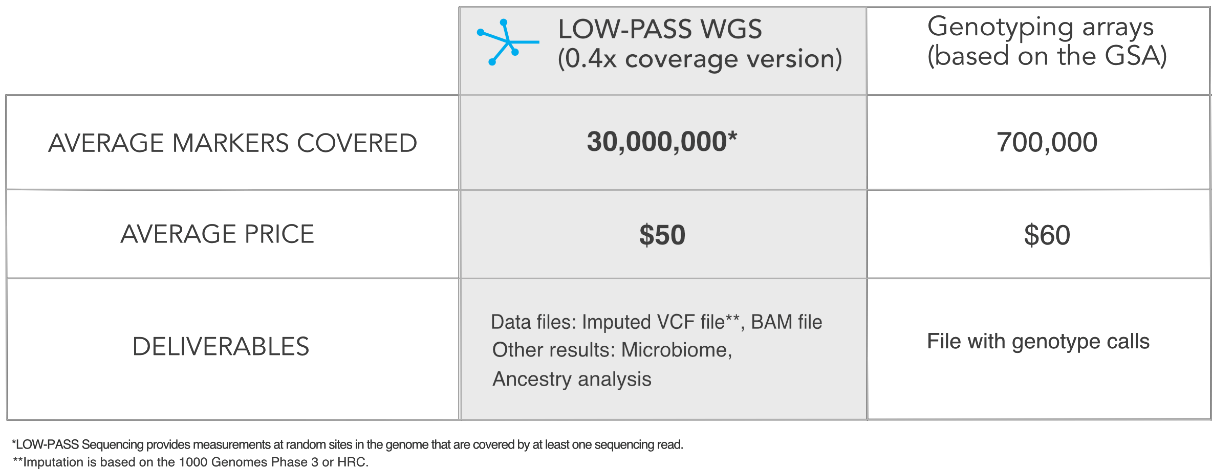
Kaja Wasik (KW): Gencove has developed a new, low-coverage sequencing technology to replace genotyping arrays. It increases the power and scale of genomics studies - at lower cost - and it allows for the discovery of new variants. This technology is an essential part of the Gencove platform, which we are using to create a genomics database that can be used for personalized medicine.
Currently, one of the biggest pain points of precision medicine is the fact that the field of genomics is still immature and the majority of diseases cannot be predicted by simply looking at somebody’s genome. To put it simply, interpreting a genome is for the most part, still like reading tea leaves.
Moreover, genomics remains inaccessible to patients due to relatively high costs. In order to use genomics in the clinic, millions more people will have to be sequenced to build up reliable genotype-phenotype databases.
We believe the creation of a robust genomics database will require active participation of the users! It was hard to find out about the quality of a local business before millions of people volunteered their reviews and made that information public on sites like Yelp. We can similarly crowdsource genomic data and corresponding phenotypes. To incentivize the collection of this data, Gencove offers affordable genome sequencing using our platform and genomic interpretation tools for Gencove or other genomic data. In a nutshell, to attract consumers, we build inexpensive and enjoyable products around genomic
applications like ancestry analysis, relative matching, and polygenic trait scores. We use the anonymized genomic data to find (and help others find) the genetic variants that really matter. Based on these variants, we build the next generation of genetic tests (precision medicine). We plan to iterate on this until there’s nothing left to discover.
F: What differentiates Gencove’s products and scientific approach from other personal genomics companies like 23andMe and AncestryDNA?
KW: Gencove’s approach relies on a novel technology – low coverage sequencing. Most direct-to-consumer genomic companies use genotyping microarrays, which measure hundreds of thousands to millions of genetic variants in a single person. It’s still less than 0.1% of the genome, and the same locations are assessed in every person, essentially leaving researchers blind to the large stretches of the genome between those measured variants. Instead, we are sequencing a random
~20% of each individual’s genome and computationally predicting the rest based on public and internal databases. Once we analyze the aggregated data across many individuals, we have the power to discover entirely new variants. In this context sequencing outperforms microarrays and costs less.
We recently opened this technology up to software developers, researchers, or anyone else via our site and our API - think Stripe for genomics. With Gencove, you can start a genomics research study in one day. We’ll handle the back-end and all of the logistics. In addition to helping researchers enroll and collect data from new individuals, our platform provides a method to import data from individuals who have already had their genetic data assayed by another company (23andMe, AncestryDNA, MyHeritage, etc). Gencove will enable you to easily combine and access all of that information in one place - independent of the file format and sequencing technology used.
F: What are some of the difficulties that you faced when implementing low- coverage sequencing in your lab?
KW: One of the key challenges we faced when turning low-coverage sequencing into a product was variation in genome coverage —
that is, when aiming for 4 million reads from each of 96, 192, or 384 samples sequenced in one sequencing run, it’s easy to end up with huge levels of variation across samples. To address this, we focused on extreme precision, in terms of input (genomic DNA) and output (genomic library) measurement and normalization. This was not easy because our protocols have been miniaturized - the majority of lab automation devices can’t handle sub microliter volumes. The MANTIS® Liquid Handler was the only piece of equipment that could reliably work with these volumes. As such, it has become an essential part of our process. The result is that almost all of our samples fall into a narrow band of coverage (below), dramatically reducing the number of samples that need to be re-sequenced to reach specification.
F: How did you go about developing a library preparation protocol that is efficient enough that you can charge $59.99 for personalized sequencing, but reproducible enough to generate trustworthy data?
KW: At Gencove, together with the New York Genome Center’s Innovation Lab, we devoted considerable resources to create the perfect sequencing library preparation protocol. We didn’t want to reinvent the wheel, so we optimized several library preparation chemistries (Illumina Nextera and Kapa Hyper Prep) for low-coverage sequencing. We introduced small changes to the protocol to make it work for us and miniaturized it to make it much cheaper. Once the library preparation was reliable enough, we automated the process to scale it up and make sure no human errors creep in. It was essential to us that the number of human steps in our library preparation protocol be minimal, so we rely on automation pretty heavily. Currently, in our most common setup we use the Opentrons, FORMULATRIX MANTIS Liquid Handler, and the Agilent Bravo to run our protocol. With this type of setup a single person can process hundreds of samples in a minimal period of time.
